We are interested in the roles that chromatin and epigenetic regulation play in development and human disease. Our studies are highly collaborative and we work with students and post doctoral fellows in the labs of Will Greenleaf, Christina Curtis, Calvin Kuo, Alistair Boettinger, Rob Malenka and Capucine van Rechem at Stanford as well as several labs at other universities. These collaborations greatly enrich the educational and intellectual environment of our lab.
We have been predominately interested in the mechanisms and function of ATP-dependent chromatin regulators in the development of the nervous system and diseases such as cancer, autism and intellectual disability. Several years ago we found that as cells progress from pluripotent stem cells to neural progenitors to committed neurons they undergo a switch (illustrated below) in chromatin regulatory mechanisms that underlie epigenetic states in the developing nervous system. This developmental switch requires a switch in the subunit composition of an ATP-dependent chromatin regulator called mSWI/SNF or BAF. We found that these complexes are made up of at least 15 subunits that are combinatorially assembled from the products of 29 genes to produce a diversity of complexes in each cell. From single cell RNA seq and immunofluorescence studies it appears that every cell has several hundred assemblies of these complexes. We envision that the subunit genes impart specificity to the complexes in the way that letters are combined into words (Wu, Lessard and Crabtree Cell 2009). To date the most informative chromatin remodeling “words” are found in pluriptotent cells, neural stem cells and committed neurons as indicated below.
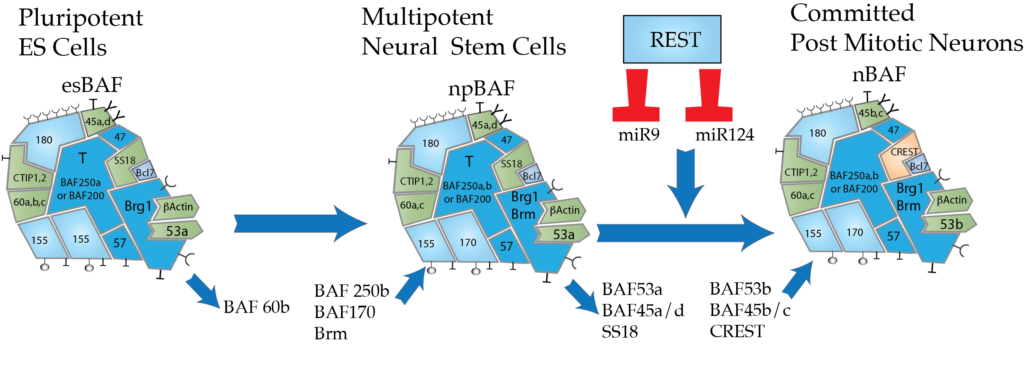 ATP-dependent chromatin remodeling in the development of the nervous system.
ATP-dependent chromatin remodeling in the development of the nervous system.
Pluripotent embryonic stem cells express a specific assembly of subunits (esBAF) and as cells enter the neural lineage a second assembly is found in neural stem cells (npBAF) which contain BAF45a/53a, SS18, and BAF53a (green below in the sub ventricular (SVZ) stem cell niche). Finally at mitotic exit three subunits are removed from this complex and replaced by three homologous subunits (BAF53b, BAF45b and CREST) to generate the nBAF complex in post mitotic neurons. These complexes contain mutually exclusive subunits at each site in the complexes. Each of these assemblies appear to be essential for the specific state of development. Andrew Yoo and Brett Staahl in our lab have found that the npBAF to nBAF switch is mediated by miR124 and 9* (Nature 2009; Nature 2011). Forcing this switch in human fibroblasts converts them to basal human neurons that then can be directed to different types of neurons (Yoo et al Nature 2011). We hope to understand how these switches are regulated during development. Just recently, we have found that the transition from npBAF to nBAF acts downstream of Wnt signaling and cellular growth factors to cause neural stem cells to exit from the cell cycle (Braun et al Genes and Development 2021).
As shown above BAF complexes have a specific assembly in neurons. Recent genomic studies have found that BAF250b or Arid1B subunit of BAF complexes is the most frequently mutated gene (specifically) in intellectual disability. Remarkably, BAF complexes are also found to have genetic variants associated with exceptional intellectual ability. The later observation suggests that BAF complexes might have played a role in the evolution of the human brain and intellect.
BAF complex subunits are also frequently mutated in autism. Recently Wendy Wenderski, a Developmental Biology student has found that mutations in the BAF53b subunit (also called ACTL6B) produce Mendelian recessive autism (Wenderski et al PNAS 2020). This is important because most genes that are associated with autism do not have a causal association and function within a specific genetic context. This genetic variability makes it difficult to conduct biochemical experiments to define the underlying mechanisms. Wendy has found that this mutation is a robust murine model of autism, independent of genetic background and even gender. We have collaborated with Rob Malenka’s and Joe Glesson’s labs to understand the way that these subunits contribute to human social behavior. Surprisingly BAF53b executes its function in adult mice and appears to act within the dorsal raphe. Recently, we have collaborated to Rob Malenka’s lab to show that serotonin receptor agonist can rapidly reverse autistic behaviors in murine models of autism (in press 2021).
ATP-Dependent Chromatin Remodeling in Human Cancer
We discovered that the genes encoding BAF complex subunit are mutated in over 20% of all human malignancies and that it plays predominantly a tumor suppressive role (Dunicef et al Cell 1994;Kadoch et al Cell 2013, Nature Genetics 2017). One of our major objectives is understanding how BAF participates in tumor suppression and to use this information to design new treatments. Diana Hargreaves and Emily Dykhuizen discovered that one aspect of BAF’s tumor protective role is to provide accessibility for the actions of TopoIIa (Nature 2013) to permit decatenation at mitosis. In the absence of BAF’s function chromosomes are not decatenated and apparently break upon cell division leading to a cascade of DNA damage and chromosomal disruption. These findings lead us to ask how BAF produces accessibility. Cigall Kadoch and Ben Stanton found that BAF rapidly evicts polycomb repressive complexes (within 5 minutes of arriving at a locus) and provides accessibility within 15 to 30 minutes (Nature Genetics 2017). These studies indicate that BAF protects cells from malignant transformation by evicting polycomb repressive complexes, producing accessibility and allowing TopoII to decatenate chromosomes at mitosis.
We are also developing new techniques to allow the in vivo understanding of epigenetic mechanisms (Hathaway et al Cell 2012, Braun et al Nature Comm 2017). These studies allow the investigation of fundamental mechanisms of epigenetic regulators in virtually any cell type at any time in development.
